The candidates for spider mite silk proteins were salivary proteins -Two functions were discovered: silk coating and fixing the mouthparts-
October 21, 2025
Research Outline
An international research group at Tokyo University of Agriculture and Technology (TUAT) has discovered that proteins previously thought to be silk proteins in the agricultural pest spider mite (Tetranychus urticae Koch) are actually salivary proteins. The team consists of Ms. Yuka Arai, a Ph.D. candidate (at the time of the research) at the Graduate School of Bio-Applications and Systems Engineering (BASE); Mr. Naoki Takeda, a Ph.D. candidate at BASE and JSPS Research Fellow DC1 (at the time of the research); Prof. Hisashi Murakami of the Division of Applied Chemistry, Institute of Engineering; Dr. Dagmar Voigt and Prof. Takeshi Suzuki of the Institute of Global Innovation Research (GIR).
Spider mites produce nanoscale silk fibers from their second appendages near their mouthparts, using them for nest-building and ballooning (dispersal by wind). Previously, proteins Fibroin-1 and Fibroin-2 were detected from the silk proteome in T. urticae and were considered strong candidates for the main components of the silk fibers (see press release dated April 21, 2021).
In this study, the research group first discovered a new protein, sFibroin-1, with a structure highly similar to Fibroin-1. They then determined that all three candidate silk proteins are produced in the salivary glands and secreted externally as salivary proteins. Furthermore, these salivary proteins were shown to potentially function both to coat the silk fiber surface and enhance its adhesion, and to be secreted onto leaf surfaces during feeding to stabilize the mouthparts. As these functions are crucial for spider mite survival, the three salivary proteins are expected to be new targets for spider mite control.

The paper was accepted for publication in Communications Biology on September 12, 2025, and was published online on October 17.
Paper title: Dual roles of salivary proteins in feeding and silk fiber coating in the spider mite Tetranychus urticae
DOI: 10.1038/s42003-025-08885-0
URL: https://www.nature.com/articles/s42003-025-08885-0
Research Background
The two-spotted spider mite (Tetranychus urticae Koch) belongs to the subphylum Chelicerata, the second most diverse group of arthropods, and is a cosmopolitan species distributed almost worldwide. This species can feed on over 1,100 plant species and is a significant agricultural pest known for its remarkable development of pesticide resistance. Because of its extremely broad host range and difficulty in control, it is referred to as a “super pest.” Given this significance, the complete genome sequence of this species was determined in 2011, marking the first such achievement for a chelicerate (Grbic et al. Nature 479, 487-492, 2011). Since then, it has been positioned as a model organism for chelicerates, and research reports utilizing its genomic information have rapidly increased.
As typified by T. urticae, mites belonging to the same subfamily Tetranychinae within the family Tetranychidae, order Trombidiformes, class Arachnida are able to produce nanoscale silk fibers. They secrete liquid silk proteins produced in the spinning glands (Note 1) from the tips of spinnerets located on the tarsal segments of the pedipalps (Note 2), the second appendages, onto leaf surfaces, spinning the silk as they move. This silk serves as a multifunctional nanobiomaterial crucial for mite survival. It is used for nest building and ballooning (dispersal by wind), functions as a cleaning tool within nests, provides a substrate for intraspecific chemical communication, and acts as a lifeline preventing falls from host plants.
However, controversy has persisted regarding the identity of the silk protein. There were 17 silk candidate genes predicted from the T. urticae genome, based on characteristic amino acid sequences. Meanwhile, a research group including Prof. Suzuki conducted silk proteome analysis (Note 3) and discovered two silk protein candidates (Fibroin-1 and Fibroin-2) distinct from the predicted 17 candidates (Arakawa et al. Journal of Proteomics 104195, 2021; see press release dated April 21, 2021). Interestingly, it had been reported in 2016 that the Fibroin-1 and Fibroin-2 genes are expressed in the salivary gland (Note 4) (Jonckheere et al. Molecular & Cellular Proteomics 15, 3594-3613, 2016). Furthermore, that report detected not only Fibroin-1 and Fibroin-2 but also sFibroin-1, which shares a highly similar structure with Fibroin-1, in the salivary proteome analysis.
The questions then arose: Are Fibroin-1, Fibroin-2, and sFibroin-1 indeed silk proteins, or are they salivary proteins? What functions do these proteins have? This study aimed to answer these questions.
Research Findings
Fluorescence in situ hybridization (FISH) (Note 5) revealed that the Fibroin-1, Fibroin-2, and sFibroin-1 genes are expressed in the dorsal podocephalic gland, one of the salivary glands, rather than in the spinning glands that produce silk proteins (Figure 1). These results suggest that these three candidate silk proteins are produced in the salivary glands and secreted externally as salivary proteins.
Silk proteome analysis using a latest mass spectrometers detected 8 of the 17 silk protein candidates predicted by Grbic et al. (2011). These proteins may function as major components of the silk fibers. Meanwhile, similar to Arakawa et al. (2021), large amounts of Fibroin-1 and Fibroin-2 were detected. Furthermore, a total of 66 salivary proteins were detected, including sFibroin-1 in addition to Fibroin-1 and Fibroin-2. Why are such large quantities and diverse types of salivary proteins detected in the silk fibers? Here, we examined two possibilities: the internal connection hypothesis (where salivary glands and spinning glands are connected internally) and the external saliva adhesion hypothesis (where saliva adheres to the silk externally).
Observation of the internal microstructure using microfocus X-ray computed tomography (µCT) (Note 6) revealed no connection between the salivary glands and the spinning glands (Figure 1). Furthermore, it was confirmed that the podocephalic canal extending from the salivary glands runs parallel to the stylet (Note 7) of the first appendage inside the rostrum and opens at the end of the rostrum. Based on these findings, the internal connection hypothesis was rejected.
Next, the team proceeded to verify the saliva adhesion hypothesis. Observations using cryo-scanning electron microscopy (cryo-SEM) (Note 8) confirmed traces of fluid patches remaining on the leaf surface near the mite's feeding site and a beads-on-a-string pattern (Note 9) of droplets strung along the silk fiber (Figure 2). Furthermore, video observation confirmed fluid secretion from the tip of the mouthparts. Based on these results, this fluid is considered to be saliva that is produced in the salivary glands, secreted externally through the podocephalic canal, and attached to the surfaces of leaves and silk fibers.
Using RNA interference (RNAi) (Note 10), suppression of Fibroin-1 and sFibroin-1 gene expression resulted in a decrease in feeding duration, feeding activity, fecundity, and survival. On the other hand, suppression of Fibroin-2 gene expression resulted in a decrease in silk filament diameter, as confirmed by analysis using atomic force microscopy (AFM) (Note 11). These results suggest that Fibroin-1 and sFibroin-1 may contribute to stabilizing feeding by secreting saliva onto the leaf surface to anchor the mouthparts when sucking leaf cell contents through the stylets. Furthermore, Fibroin-2 may coat the silk filament surface to impart adhesiveness.
This study indicates that proteins previously considered candidate silk proteins in T. urticae are actually salivary proteins, potentially performing two crucial roles: stabilizing the mouthparts during feeding and coating the silk. This represents a novel discovery of salivary function in phytophagous chelicerates. Notably, suppression of Fibroin-1 and sFibroin-1 gene expression inhibited the mite's feeding behavior and reduced both fecundity and survival. Therefore, these genes are promising targets for RNAi-based biopesticides (Note 12) with functional mechanisms distinct from conventional synthetic pesticides. Moving forward, we will advance research and development toward the practical application of RNAi-based biopesticides targeting these genes. Concurrently, we will elucidate the mechanisms of mouthparts stabilization and thread coating via saliva, and further investigate the evolution of arthropod saliva.
Research Structure
This work was conducted by an international research group consisting of Ms. Yuka Arai (completed the Ph.D. coursework in March 2025) and Dr. Naoki Takeda (completed the Ph.D. program in March 2025) at BASE, TUAT, Prof. Hisashi Murakami at the Division of Applied Chemistry, Institute of Engineering, TUAT, Prof. Takeshi Suzuki at GIR, TUAT, Dr. Shinya Komoto of the Okinawa Institute of Science and Technology, and Dr. Dagmar Voigt of the Technical University of Dresden (GIR foreign researcher at the time of the research). This work was supported in part by JSPS KAKENHI grants 16K18661, 18H02203, 21H02193, and 24K21256, as well as the Cabinet Office, Government of Japan, Moonshot Research and Development Program for Agriculture, Forestry and Fisheries (JPJ009237; funding agency: Bio-oriented Technology Research Advancement Institution.
Reference Press Release
・ "Identification of Spider Mite Silk Genes " (April 21, 2021)
Glossary
Note 1) Spinning glands
A pair of giant unicellular glands, the cytoplasm of which is filled with numerous vesicles containing granules. They are located on the ventral side of the proterosoma (consisting of the gnathosoma and propodosoma bearing legs I and II), in front of the brain (nervous mass or synganglion), and to the side of the esophagus. They penetrate the interior of the pedipalps and open at the tip of the spinneret located in the tarsus.
Note 2) Pedipalps
The second pair of appendages consisting of the tarsus, tibia, genu, femur, and trochanter from the distal to proximal end, with the coxa being integrated into the body. The tibia has a claw that grips the leaf surface during feeding. The distal end of the tarsus has a spinneret that secretes silk proteins, as well as sensory setae for olfactory and gustatory stimuli.
Note 3) Proteome analysis
A comprehensive method for analyzing protein identity and abundance.
Note 4) Salivary glands
Composed of a pair each of anterior and dorsal podocephalic glands, both located in the proterosoma and containing numerous granules. Podocephalic canals extend from these podocephalic glands to the end of the rostrum. The proterosoma also contains several other secretory glands that may function as salivary glands.
Note 5) Fluorescence in situ hybridization (FISH)
A technology that uses fluorescent RNA probes to visualize the localization of target RNA with high sensitivity. In this study, the RNAscopeTM method was used.
Note 6) Microfocus X-ray CT (µCT)
A technique for non-destructively observing the internal microstructure in 3D by irradiating a rotating sample with X-rays and reconstructing the acquired images. Also known as 3D X-ray microscopy.
Note 7) Stylet
The movable digits of the pair of chelicerae, the first appendages. They are whip-like and have a trough-like cross section, with both movable digits joined together to form the stylet. During feeding, the stylet penetrates mesophyll cells and is used to inject saliva and suck out the cellular contents that have pre-orally digested. In male-male aggression over females, the stylet is also used as a weapon to stab and kill opponents.
Note 8) Low-temperature scanning electron microscope (cryo-SEM)
A scanning electron microscope (SEM) is used to observe samples in a frozen state. SEM is a type of electron microscope that detects secondary electrons, backscattered electrons, and X-rays emitted by electron beam irradiation to observe the surface structure of a sample. Cryo-SEM allows high-resolution observation of biological samples while preserving their natural structure by rapidly freezing the sample using liquid nitrogen or other methods.
Note 9) Beads-on-a-string pattern
A structure in which fluid droplets (beads) are arranged like beads along a silk filament (string). This structure is formed by the tendency of the fluid to form into a sphere due to surface tension and by the fluid existing along the string. In this study, this was observed as morphological evidence that saliva was attached to the silk fibers.
Note 10) RNA interference (RNAi)
A phenomenon in which the expression of a target gene is suppressed by introducing double-stranded RNA (dsRNA) that is complementary to the base sequence of the gene. This technique is used to investigate gene function.
Note 11) Atomic force microscope (AFM)
A type of microscope used to observe sample surfaces at the nanoscale. An AFM is a device that brings a sharp probe tip close to the sample surface, detects the minute forces acting between the probe tip and the sample surface, and measures the shape and properties of the surface in detail. It can be used in air or liquid, and is capable of observing a wide range of samples, from metals to biological materials.
Note 12) RNAi-based biopesticides
Biopesticides that use dsRNA as their active ingredient and RNAi as their mechanism of action. They are also called RNA pesticides or RNAi pesticides. Because they act sequence-specifically, their target range can be designed with species-level resolution.
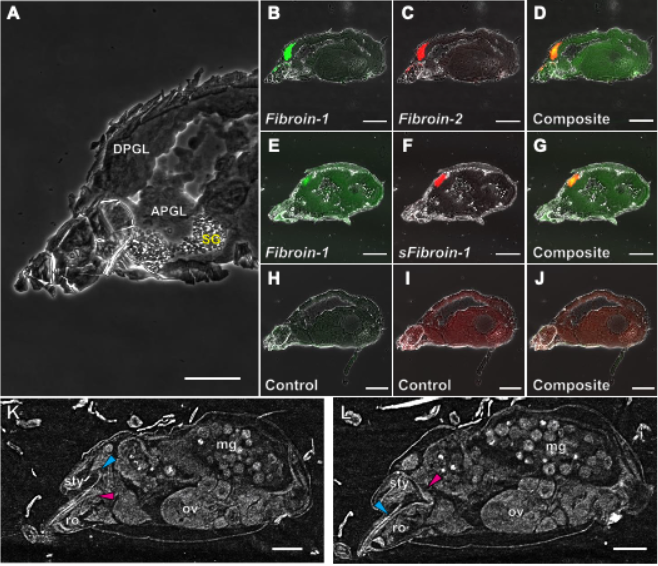
Figure 1. (A) Internal structure of the proterosoma of the two-spotted spider mite, Tetranychus urticae. (B-J) Visualization of the expression localization of Fibroin-1, Fibroin-2, and sFibroin-1 genes by FISH. (K, L) X-ray images of the internal morphology by µCT. Cyan and magenta arrowheads indicate the stylet and podocephalic canal, respectively. APGL, anterior podocephalic gland; DPGL, dorsal podocephalic gland; mg, midgut; ov, ovary; ro, rostrum; SG, spinning gland; sty, stylophore.
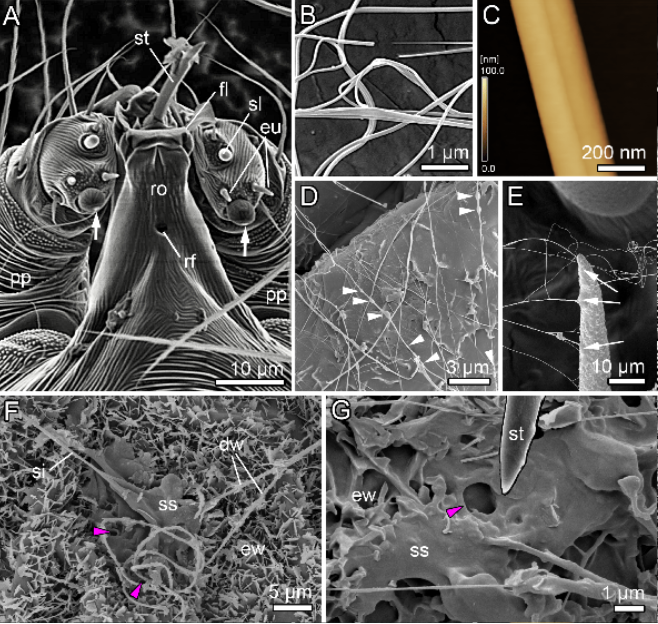
Figure 2. (A) SEM image of the gnathosoma of the two-spotted spider mite, Tetranychus urticae. (B) Cryo-SEM image and (C) AFM image of a silk fiber. (D-G) Cryo-SEM images of silk and saliva-like fluid on a plant. dw, detached wax crystal; eu, eupathidium; ew, epicuticular wax crystal; fl, flap; pp, pedipalps; rf, rostral fossette; ro, rostrum; si, silk fiber; sl, solenidion; ss, silk-saliva mixture; st, stylet.
◆Inquiries about Research◆
Tokyo University of Agriculture and Technology Graduate Institute of Global Innovation Research
Professor Takeshi Suzuki
TEL: 042-388-7278
E-mail: tszk (put @ here) cc.tuat.ac.jp
